Why?
The identification of drug-target interaction (DTI) represents a costly and time-consuming step in drug discovery and design. Computational methods capable of predicting reliable DTI play an important role in the field. Algorithms may aim to design new therapies based on a combination of approved drugs. Recently, recommendation methods relying on network-based inference in connection with knowledge coming from the specific domain have been proposed. However no knowledge base offering the drug-target prediction service through web interfaces has been reported.
DT-Hybrid Web (DT-Web) is a web-based interface to the DT-Hybrid Algorithm (Alaimo et al., 2013) which extends a well-established recommendation technique by domain-based knowledge such as drug and target similarity. The interface offers users the following features:
- browse the predictions computed to analyze the performances of the DT-Hybrid, and based on a periodically synchronized version of DrugBank;
- upload custom data on which a prediction using DT-Hybrid will be computed;
- predict combinations of drugs whose targets are at an optimal distance from some candidates disease genes.
Definitions
Below is a quick definition list to get you started.
- Small Molecule Drug
- A drug from largely synthetic origins, typically under 1000 MW
- Biotech Drug
- Drugs which are peptide, protein or nucleic acid drugs
- Target
- A protein, macromolecule, nucleic acid, or small molecule to which a given drug binds, resulting in an alteration of the normal function of the bound molecule and a desirable therapeutic effect. Drug targets are most commonly proteins such as enzymes, ion channels, and receptors.
- Enzyme
- A protein which catalyzes chemical reactions involving the a given drug (substrate). Most drugs are metabolized by the Cytochrome P450 enzymes.
- Transporter
- A membrane bound protein which shuttles ions, small molecules or macromolecules across membranes, into cells or out of cells.
- Carrier
- A secreted protein which binds to drugs, carrying them to cell transporters, where they are moved into the cell. Drug carriers may be used in drug design to increase the effectiveness of drug delivery to the target sites of pharmacological actions.
Browsing
Among the offered features, DT-Web gives an user the opportunity to see a version of the DrugBank database enriched with the predictions obtained through the application of the DT-Hybrid algorithm. The consultation of such information can be made through the appropriate section "Browse" of our website. The user can see a list of drugs, or a list of targets, or a list of drugs filtered through the categories defined in DrugBank.
In the drugs list, the user can find a table that contains key information about the drug (name, weight, 2D structure, categories and therapeutic indication). A special link will, therefore, give access to a more detailed view for each drug. The list of drugs filtered by categories differs from the full list for the possibility to select a category and view only the drugs associated with it. The information shown in the latter are exactly the same as the full list.
The detailed view for a drug consists in a page containing the main information for that drug. These include:
- The generic information that identify such a drug;
- The information related to its pharmacology;
- The information on its interactions with other drugs and foods;
- a list of its validated and predicted targets;
- a list of its enzymes;
- a list of its transporters;
- a list of its carriers.
A more detailed description of each field is available here.
| Field | Description |
|---|---|
| Name | Standard name of drug as provided by drug manufacturer. |
| Accession Number | Unique DrugBank accession number consisting of a 2 letter prefix (DB) and a 5 number suffix. |
| Type | Small molecule or Biotech. Small molecule drugs are drugs which are not from biological origin and are synthesized. Biotech drugs consist of peptide, protein or nucleic acid drugs. |
| Description | Description of the drug describing general facts, composition and/or preparation. |
| 2D structure | The 2D chemical structure. |
| MOL structure | A link to download the drug structure in MOL format. |
| Molecular Weight | Molecular weight in g/mol, determined from molecular formula or sequence. |
| Groups | Can be one or more of:
|
| Monoisotopic Weight | The sum of the masses of the atoms in a molecule using the unbound, ground-state, rest mass of the principle (most abundant) isotope for each element instead of the isotopic average mass. |
| SMILES | Isomeric SMILES string corresponding to drug structure. |
| Indication | Description or common names of diseases that the drug is used to treat. |
| Mechanism of action | Description of how the drug works or what it binds to at a molecular level. |
| Absorption | Description of how much of the drug or how readily the drug is taken up by the body. |
| Protein binding | Percentage of the drug that is bound in plasma proteins. |
| Biotransformation | Description of how the drug works at a clinical or physiological level. |
| Route of elimination | Route by which the drug is eliminated. Drugs are cleared primarily by the liver and kidneys. |
| Toxicity | Lethal dose (LD50) values from test animals, description of side effects and toxic effects seen in humans. |
| Affected organisms | Names of organisms for which the drug is most effective. |
| Drug Interactions | Drugs that are known to interact, interfere or cause adverse reactions when taken with this drug . |
| Food Interactions | Foods that are known to interact, interfere or cause adverse reactions when taken with this drug. |
| Targets | A list of all targets known to be associated with that drug. |
| Enzymes | A list of all enzymes known to be associated with that drug. |
| Transporters | A list of all transporters known to be associated with that drug. |
| Carriers | A list of all carriers known to be associated with that drug. |
The list of targets is structured as the list of drugs: a table lists the possible targets in the database. This table contains information about the name of the target and its function. By clicking on each target in the list, the user can access a page that contains key information on it: name, synonyms, function, protein and gene sequences, GO classification, and associated drugs.
Searching
DT-Web allows users to look for a specific drug by using the search field placed on the website header present in each page, or on the home page at the center of the window. Once the user enter a text, by clicking on the "Search" button (where available) or by pressing "Enter" the search process will start and a list of drugs corresponding with the one the user is looking for will be shown. When the user specifies an accession number or a name, its page will be automatically opened without displaying a list of results.
Among the keywords that an user can enter in the search field we have:
| Type | Example |
|---|---|
| Drug ID | DB00201 |
| Name | Caffeine |
| Secondary accession number | APRD00673 |
| Description | Caffeine's most notable pharmacological effect is as a central nervous system stimulant, increasing alertness and producing agitation |
Job Types
DT-Web allows user to execute two different kind of jobs on their own data: DTI Prediction, Drug Combination. The first one allows an user to upload his custom data on which a prediction using DT-Hybrid will be computed, while the second one can be used to predict combinations of drugs whose targets are at an optimal distance from the candidates disease genes (indirect targets) to reduce drug side effects.
DTI Prediction
In this type of job, the DT-Hybrid algorithm is applied to the data submitted by the user to compute a list of predictions. For each drug in such list, the algorithm associates a score that indicates how much the predicted targets are correlated with the original ones, and a p-value that indicates the probability of obtaining such score by chance. Among all the predictions for each drug, the ones that minimize the p-value are selected and returned to the user. To estimate the correlation between the predicted targets and the validated ones, each target is annotated with GO terms. For each pair of terms, then, a similarity measure is calculated based on the distance in the ontology graph. Given, then, a set of predicted targets for a drug, the correlation with the validated ones corresponds to the minimum similarity between the possible pairs of ontological terms associated with each target.
The result of this procedure is displayed both in the form of table in which all the information are explained, and in the form of a graph in which a user can quickly see the new structure of the DTI network after applying the methodology.
The input of the job consists of 3 text file:
- one containing the description of the original DTI network;
- one containing the similarity between drugs in the network (optional);
- one containing the similarity between targets in the network (optional).
In the first file, each row describes a drug and its target. For example:
DRUG 1 TARGET 1,TARGET 2 DRUG 2 TARGET 2,TARGET 3 DRUG 3 TARGET 2
The other two files contain in each row a value of similarity (ranging from 0 to 1) associated with a pair of drugs (targets). For example:
target.similarity.txt TARGET 1 TARGET 2 0.5 TARGET 1 TARGET 3 0.3 TARGET 2 TARGET 3 0.8 drug.similarity.txt DRUG 1 DRUG 2 0.6 DRUG 1 DRUG 3 0.3 DRUG 2 DRUG 3 0.9
The user can also specify the parameters of the DT-Hybrid algorithm. For more information about them, refer to Alaimo et al. 2013.
Drug Combination
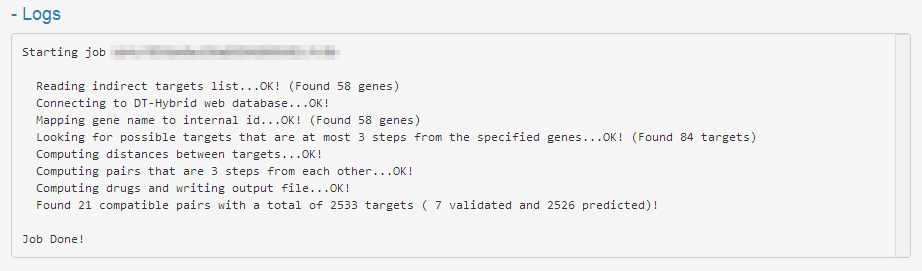
In this type of job, the user specifies in a text box, a list of genes (Gene Names). The algorithm will track down all the drugs that may have an indirect influence on them based on their position in some pathway. Starting from such list of genes, the algorithm will compute all possible pairs of targets (reported in drugbank or predicted by DT-Hybrid) which are at most 3-4 steps away from the input genes. Such 3-4 distance value in the literature is claimed to reduce drug side effects. However this default parameter can be modified by the user. Finally, all drugs targeting such pairs are extracted and returned as combination prediction.
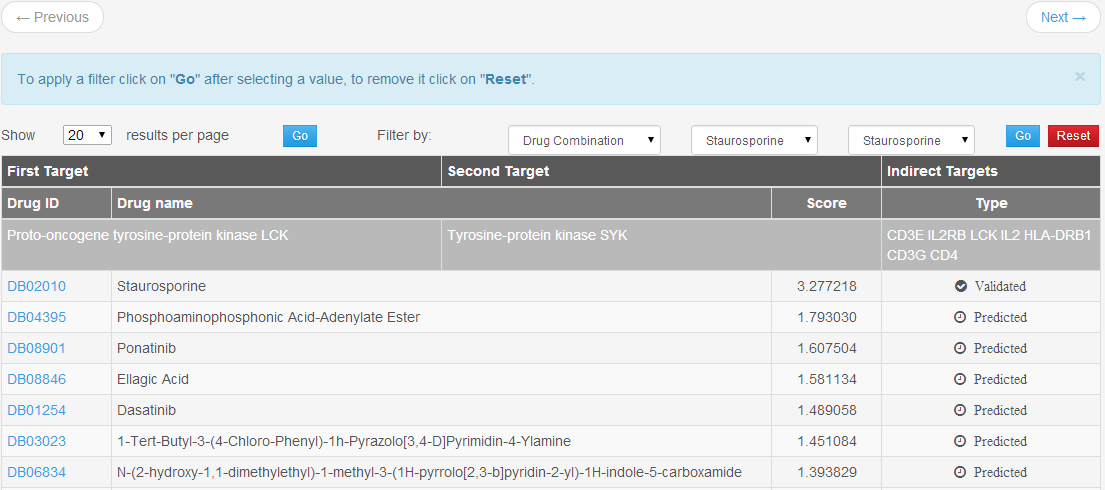
The result of this procedure is displayed in the form of table in which all predictions are exposed. The user can also filter the results in this table by drug combination or according to the intermediate target found in some pathway or according to the type of interaction (whether it is validated or predicted).
Submitting a job
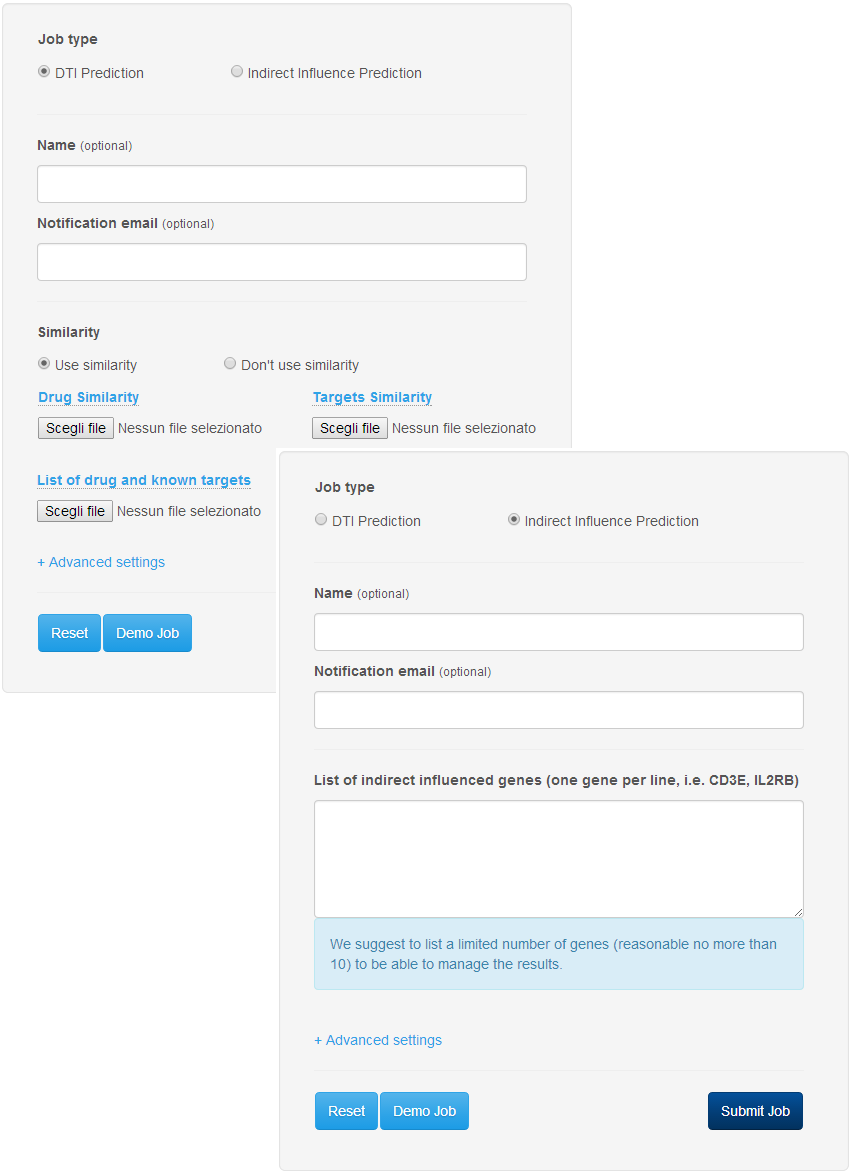
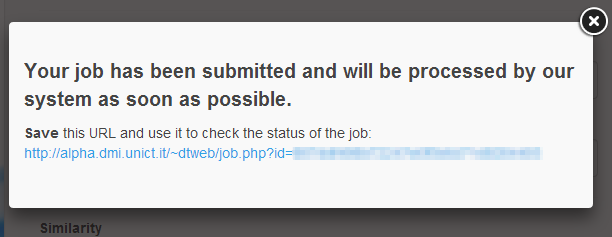
The procedure for the submission of a job is extremely simple. To access the submission page just click on the button "Submit your job" in the header of the DT-Web website. In the submission page (Figure 1), the user can select the type of job (DTI prediction or Indirect Target prediction), give a name to the job so it can be easily identified, and specify an email address to receive notifications about the status of the job. After selecting the type, the user can choose to enter the data needed to start a computation, or, by using the "Demo Job" button, load sample data to start an example of computation. The user can finally click on "Submit Job" button to add the job to the computation queue. If the procedure is successful, the user will be notified with an appropriate message on the screen which contains a link that can be stored for later access to the job (Figure 2).
All computations are asynchronous and are carried out as soon as a computation slot becomes free. The time required for the completion of a job depends on the amount of data sent and the number of other jobs in the queue. If the user has specified an email address, he will be notified upon completion of the job.
Consulting the results of a job
After the submission of a job, a URL will appear in the screen that the user can use to check the status of a job and view the results when ready. The status page is highly dependent on the type of the selected job, but one can identify four main sections:
- General information (Figure 3);
- Logs (Figure 4);
- Results of the job (Figure 5 and 6);
- Secondary Actions (Figure 7 and 8).
The first section (Figure 3) displays general information about the job: its type ,the internal ID, its current status, the date of submission, start of computation, and end of computation.
The possible states are as follows:
- QUEUED: The job is waiting for a free computation slot;
- PENDING: the job is running and the data is being processed;
- DONE: the job has finished successfully and the results are ready to be displayed;
- ERROR: the job has finished due to an error, and the results were not generated.
The Log section (Figure 4) allows you to view all information on the progress of a computation and can be helpful to identify the reasons why something went wrong during the execution of the job.
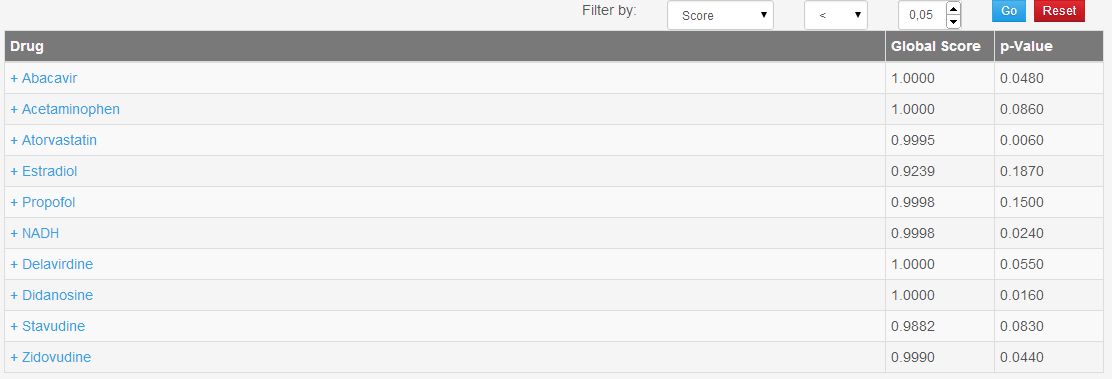
The results section for a DTI prediction job (Figure 5) consists of a table in which all the drugs specified by the user are shown with the correlation scores and p-values. The user, by clicking on a drug, can view its list of predictions associated with their score computed by DT-Hybrid. Finally, the user can choose to filter the results by using the appropriate boxes placed on top of the table.
The results section for a Indirect Target prediction job (Figure 6) provides a list of drug associated with a pair of targets, calculated using the procedure described above, and the type of binding (validated or predicted). The user can choose, by using the buttons at the top of the table, to filter associations by drug combination, or target, or by type.
For both types of jobs an user can download the predictions using the appropriate button (Figure 7 and 8) and remove the job from the server.
For a DTI prediction job (Figure 7) the user can download the user-supplied datasets and graphically display the results. The graphical visualization consists in a graph of the DT interactions in which all the predictions were added. This graph is displayed through the Cytoscape Web interface and for that reason it requires the installation of the Flash plugin in your browser.